require(Yano)Loading required package: Yano── Attaching packages ────────────────────────────────────────────── Yano 1.2 ──
✔ dplyr 1.1.4 ✔ Seurat 5.3.0
✔ ggplot2 4.0.0 exp.1 <- ReadPISA("./visium/section1/Anterior/exp/", prefix = "sec1_ant_")
image.1 <- Read10X_Image("./visium/section1/Anterior/spatial/")
new.names <- paste0("sec1_ant_",Cells(image.1))
image.1 <- RenameCells(image.1, new.names = new.names)
## QuickRecipe() is actually an integration function of Seurat workflow
obj.1 <- QuickRecipe(exp.1[, Cells(image.1)], verbose = FALSE)
obj.1[['slice1']] <- image.1[colnames(obj.1),]
obj.1$orig.ident <- "sec1_ant"
exp.2 <- ReadPISA("./visium/section1/Posterior/exp/", prefix = "sec1_pos_")
image.2 <- Read10X_Image("./visium/section1/Posterior/spatial/")
new.names <- paste0("sec1_pos_",Cells(image.2))
image.2 <- RenameCells(image.2, new.names = new.names)
obj.2 <- QuickRecipe(exp.2[,Cells(image.2)], verbose = FALSE)
obj.2[['slice2']] <- image.2[colnames(obj.2),]
obj.2$orig.ident <- "sec1_pos"
exp.3 <- ReadPISA("./visium/section2/Anterior/exp/", prefix="sec2_ant_")
image.3 <- Read10X_Image("./visium/section2/Anterior/spatial/")
new.names <- paste0("sec2_ant_",Cells(image.3))
image.3 <- RenameCells(image.3, new.names = new.names)
obj.3 <- QuickRecipe(exp.3[, Cells(image.3)], verbose = FALSE)
obj.3[['slice3']] <- image.3[colnames(obj.3),]
obj.3$orig.ident <- "sec2_ant"
exp.4 <- ReadPISA("./visium/section2/Posterior/exp/", prefix="sec2_pos_")
image.4 <- Read10X_Image("./visium/section2/Posterior/spatial/")
new.names <- paste0("sec2_pos_",Cells(image.4))
image.4 <- RenameCells(image.4, new.names = new.names)
obj.4 <- QuickRecipe(exp.4[, Cells(image.4)], verbose = FALSE)
obj.4[['slice4']] <- image.4[colnames(obj.4),]
obj.4$orig.ident <- "sec2_pos"
# Merge all processed Seurat object
obj <- merge(obj.1, y = list(obj.2, obj.3, obj.4))
obj <- QuickRecipe(obj)Set default assay to RNA
object <- NormalizeData(object, normalization.method = "LogNormalize", scale.factor = 10000)
Normalizing layer: counts.1
Normalizing layer: counts.2
Normalizing layer: counts.3
Normalizing layer: counts.4
object <- FindVariableFeatures(object, selection.method = "vst", nfeatures = 3000)
Finding variable features for layer counts.1
Finding variable features for layer counts.2
Finding variable features for layer counts.3
Finding variable features for layer counts.4
object <- ScaleData(object, features = @features)
Centering and scaling data matrix
object <- RunPCA(object, features = @features
PC_ 1
Positive: Trf, Apod, Plekhb1, Mbp, Cnp, Plp1, Mobp, Car2, Mag, Dbi
Cryab, Mog, Cldn11, Bcas1, Sept4, Mal, Ndrg1, Ptgds, Gatm, Ermn
Qdpr, Ppp1r14a, Sparc, Gsn, Ugt8a, Gfap, Fa2h, Gm20425, Pllp, Tmem88b
Negative: Calm2, Ctxn1, Rtn1, Chn1, Nrgn, Syt1, Ppp3r1, Cx3cl1, Ptprn, Gria2
Gpm6a, Sept5, Ptk2b, Gp1bb, Enc1, Calm1, Fbxl16, Basp1, Arf3, Snca
Meg3, Camkv, Hpcal4, Syt5, Chst1, Nsg2, Icam5, Ppp3ca, Pld3, Dclk1
PC_ 2
Positive: Tmsb4x, Nrgn, Inf2, Ctxn1, Ngef, Gpr88, Gng7, Ddn, Adcy5, Pde1b
Camkv, Rgs9, Phactr1, Kcnip2, Klhl2, Ptk2b, Lrrc10b, Ptpn5, Tac1, Cx3cl1
Adora2a, Bcl11b, Efnb3, Crip2, Mal, Pde2a, Ppp1r1a, Tmem158, Basp1, Hpca
Negative: Cbln1, Car8, Neurod1, Pcp2, Gm2694, Ppp1r17, Lhx1os, Zic1, Nrep, Shf
Rgs8, Kcnc3, Grin2c, Ank1, Inpp5a, Homer3, Grm4, Pvalb, Kcnc1, Il16
Sphkap, Icmt, Calb2, Trim62, Gng13, Il20rb, Chn2, Pagr1a, Dusp5, Atp2a3
PC_ 3
Positive: Cck, Stmn1, Gnas, Nrn1, 3110035E14Rik, Slc17a7, Olfm1, Vsnl1, Tbr1, Lingo1
Stx1a, Slc30a3, Ncald, Dkk3, Efhd2, Rtn4r, Neurod6, Miat, 1110008P14Rik, Gm11549
Mpped1, Basp1, Pde1a, Gabbr2, Nsmf, Fxyd7, Syt13, Mical2, E130012A19Rik, Osbpl1a
Negative: Penk, Rgs9, Adora2a, Pde10a, Ppp1r1b, Gpr88, Drd1, Syndig1l, Gpr6, Tac1
Lrrc10b, Rxrg, Adcy5, Scn4b, Pde1b, Rasd2, Gng7, Rarb, Slc35d3, Tmem158
Actn2, Serpina9, Strip2, Slc32a1, Ankrd63, Kcnab1, Dmkn, Gnal, Pcp4l1, St8sia3
PC_ 4
Positive: Scn4b, Mbp, Slc24a2, Qdpr, Mobp, Mag, Bcas1, Nefm, Plp1, Mal
Plekhb1, Vamp1, Nefl, Sept4, Ermn, Tspan2, Mog, Arpp19, Trf, Cnp
Cryab, AI593442, Pex5l, Tmem88b, Gpr37, Ppp1r14a, Pllp, Uchl1, Kcnab1, Snap25
Negative: Igf2, Islr, Igfbp2, Slc13a4, Mgp, Fmod, Slc6a20a, Aebp1, Ogn, Nnat
Slc6a13, Col1a2, Fabp7, Aldh1a2, Rbp1, Gjb2, Pcolce, Slc22a6, Slc13a3, Bmp7
Dcn, Fn1, Serping1, Col1a1, Crabp2, Ifitm3, Efemp1, Bgn, Vim, Fbln1
PC_ 5
Positive: Itpr1, Itpka, Zbtb18, Cplx2, Adcy1, Rnf112, Nptx1, Lmo4, Hpca, Neurod2
Dkk3, Sptbn2, Camk4, Cnr1, Cnksr2, Arpp19, Fbxl16, Tmem132a, Igf2, Prkcb
Atp1a1, Dgkz, Lzts3, Mmp17, Ppp1r1b, Neurl1a, Slc8a2, Gas7, Slc13a4, Fam212b
Negative: Slc6a11, Gng4, Nrip3, Scg2, Ptpro, Sp8, Dlx1, Hap1, Cdhr1, Th
Doc2g, Zcchc12, Pcbp3, Pbx3, Scgn, Dcx, Tshz1, Tuba1a, A230065H16Rik, Tubb3
Cacng5, Pcp4l1, Cpne4, Lgr5, Lgr6, Sp9, Dlx2, Tmem130, Adamts19, Tubb2a
object <- FindNeighbors(object, dims = 1:20)
Computing nearest neighbor graph
Computing SNN
object <- FindClusters(object, resolution = 0.5)Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
Number of nodes: 12148
Number of edges: 388028
Running Louvain algorithm...
Maximum modularity in 10 random starts: 0.9315
Number of communities: 16
Elapsed time: 1 secondsobject <- RunUMAP(object, dims = 1:20)
14:56:12 UMAP embedding parameters a = 0.9922 b = 1.112
14:56:12 Read 12148 rows and found 20 numeric columns
14:56:12 Using Annoy for neighbor search, n_neighbors = 30
14:56:12 Building Annoy index with metric = cosine, n_trees = 50
0% 10 20 30 40 50 60 70 80 90 100%
[----|----|----|----|----|----|----|----|----|----|
**************************************************|
14:56:13 Writing NN index file to temp file /local/tmp/RtmpTOUqEJ/file45e9830efc0
14:56:13 Searching Annoy index using 1 thread, search_k = 3000
14:56:17 Annoy recall = 100%
14:56:17 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
14:56:18 Initializing from normalized Laplacian + noise (using RSpectra)
14:56:19 Commencing optimization for 200 epochs, with 500526 positive edges
14:56:19 Using rng type: pcg
14:56:24 Optimization finishedp1 <- DimPlot(obj, group.by = "orig.ident")
p2 <- DimPlot(obj)
p1 + p2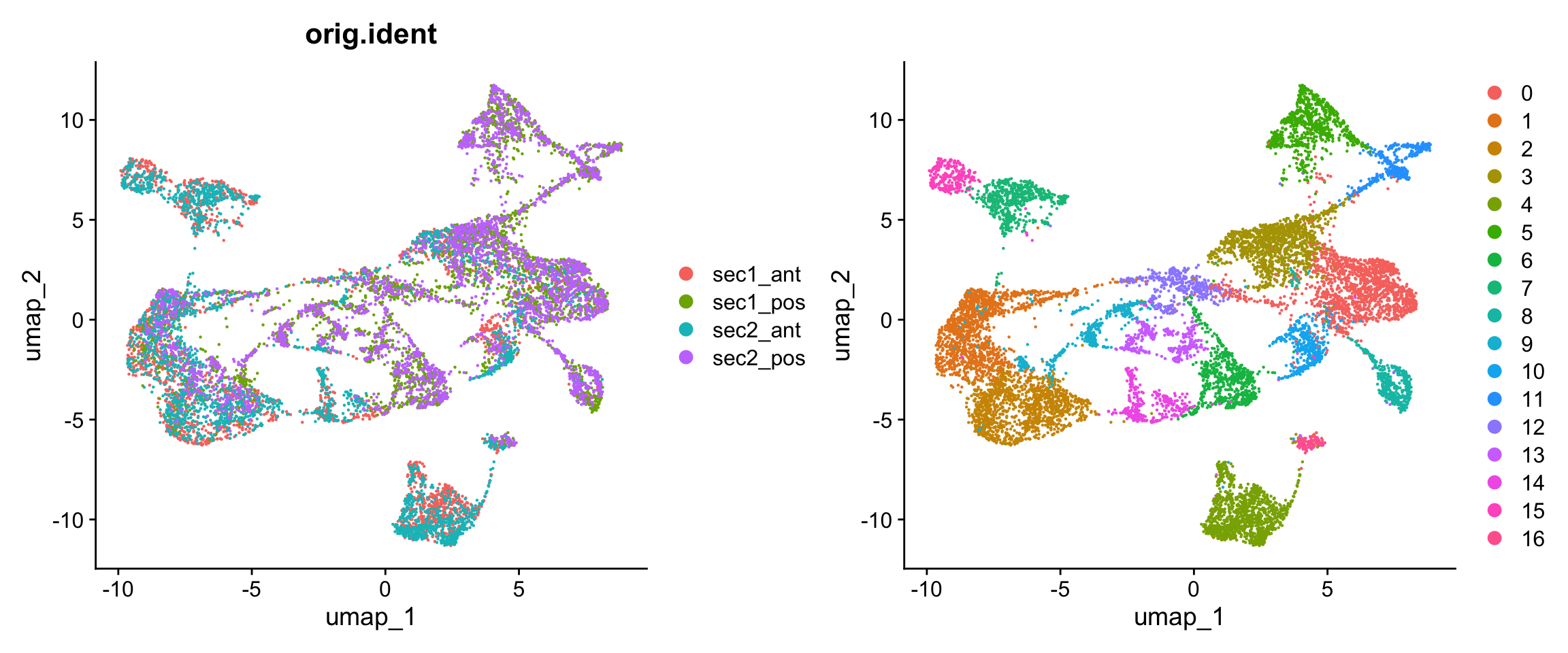
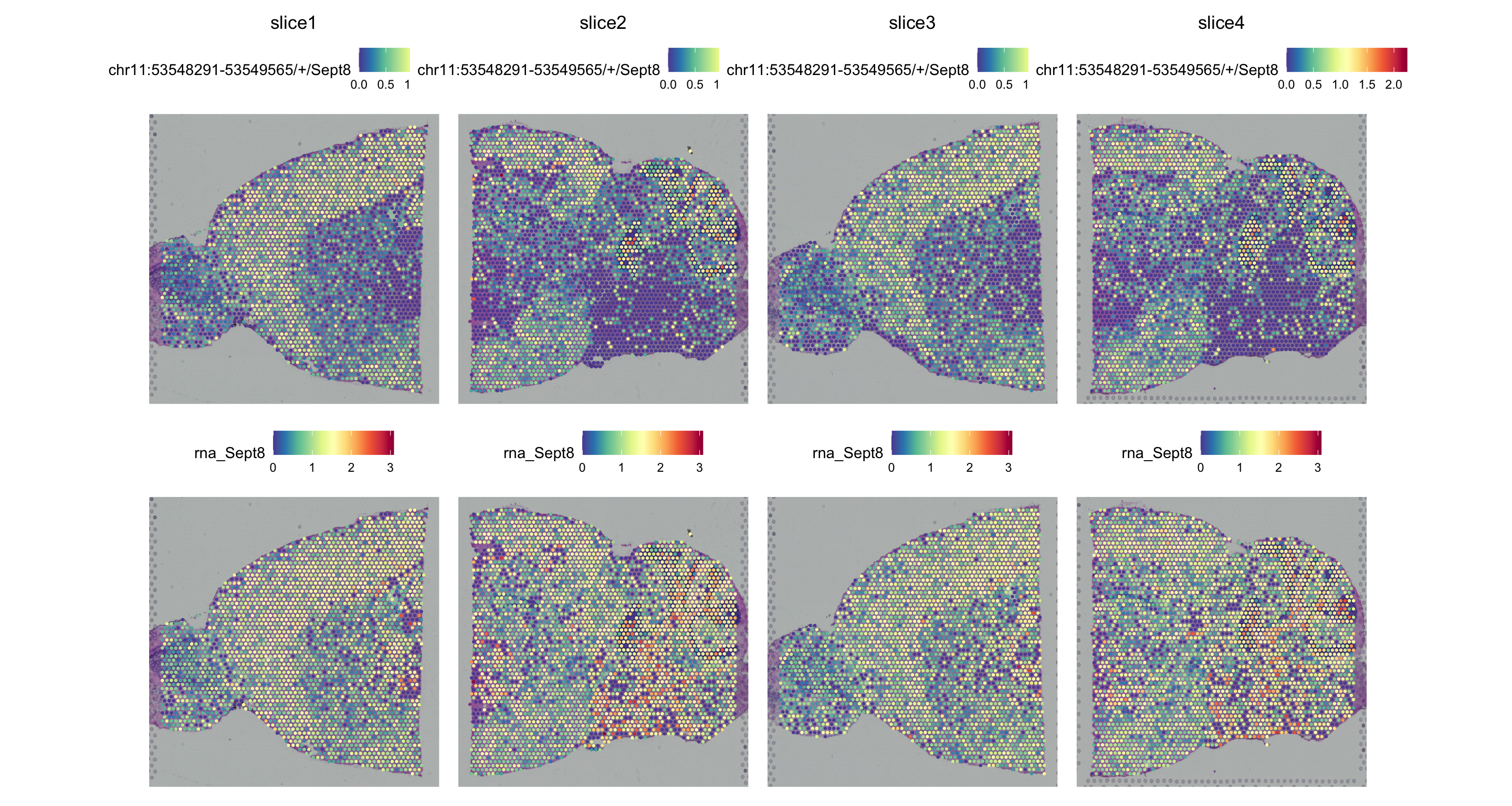
SpatialPlot(obj, pt.size.factor = 2) 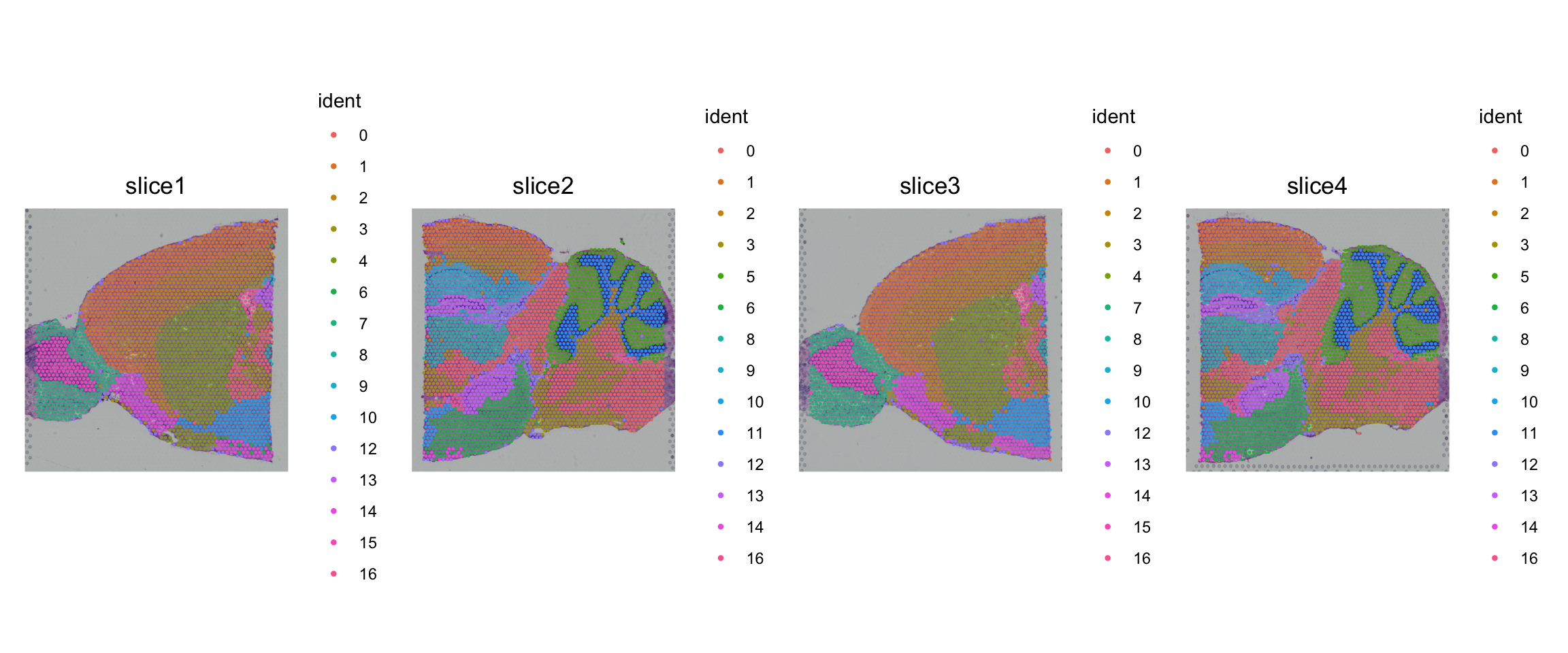
exon.1 <- ReadPISA("./visium/section1/Anterior/exon/", prefix = "sec1_ant_", cells = Cells(obj))
exon.2 <- ReadPISA("./visium/section1/Posterior/exon/", prefix = "sec1_pos_", cells = Cells(obj))
exon.3 <- ReadPISA("./visium/section2/Anterior/exon/", prefix = "sec2_ant_", cells = Cells(obj))
exon.4 <- ReadPISA("./visium/section2/Posterior/exon/", prefix = "sec2_pos_", cells = Cells(obj))
exon <- mergeMatrix(exon.1, exon.2, exon.3, exon.4)'as(<dgTMatrix>, "dgCMatrix")' is deprecated.
Use 'as(., "CsparseMatrix")' instead.
See help("Deprecated") and help("Matrix-deprecated").obj[['exon']] <- CreateAssayObject(exon[, Cells(obj)], min.cells = 20)
DefaultAssay(obj) <- "exon"
obj <- NormalizeData(obj)
obj <- ParseExonName(obj)Working on assay exonobj <- RunAutoCorr(obj)Working on assay : exon
Run autocorrelation test for 80178 features.
Runtime : 32.13648 secs
38784 autocorrelated features.obj <- RunSDT(obj, bind.name = "gene_name", bind.assay = "RNA")Working on assay exon.
Working on binding assay RNA.
Use predefined weight matrix "pca_wm".
Processing 38784 features.
Processing 15696 binding features.
Retrieve binding data from assay RNA.
Use "data" layer for test features and binding features.
Using 63 threads.
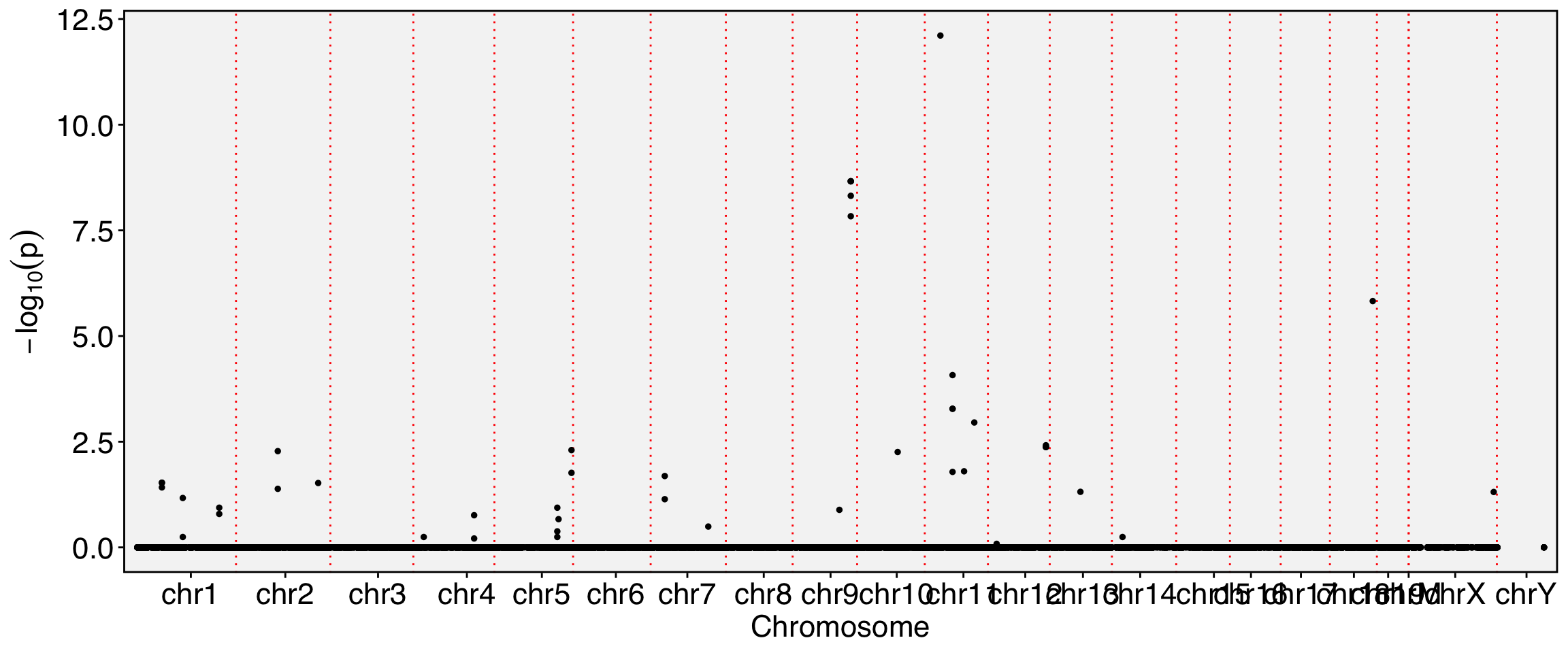
Runtime : 11.01335 mins.FbtPlot(obj, val = "gene_name.padj")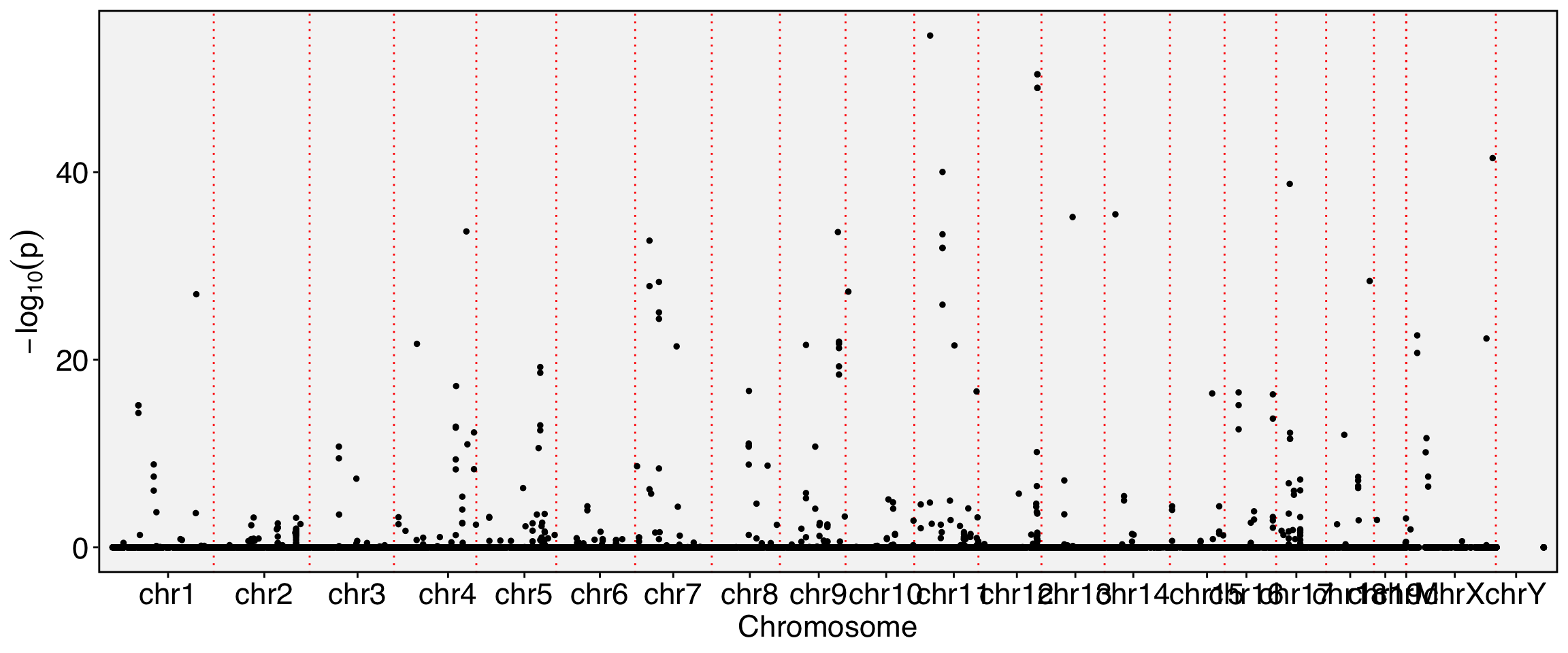
obj[['exon']][[]] %>% filter(gene_name.pval<1e-40) chr start end gene_name strand
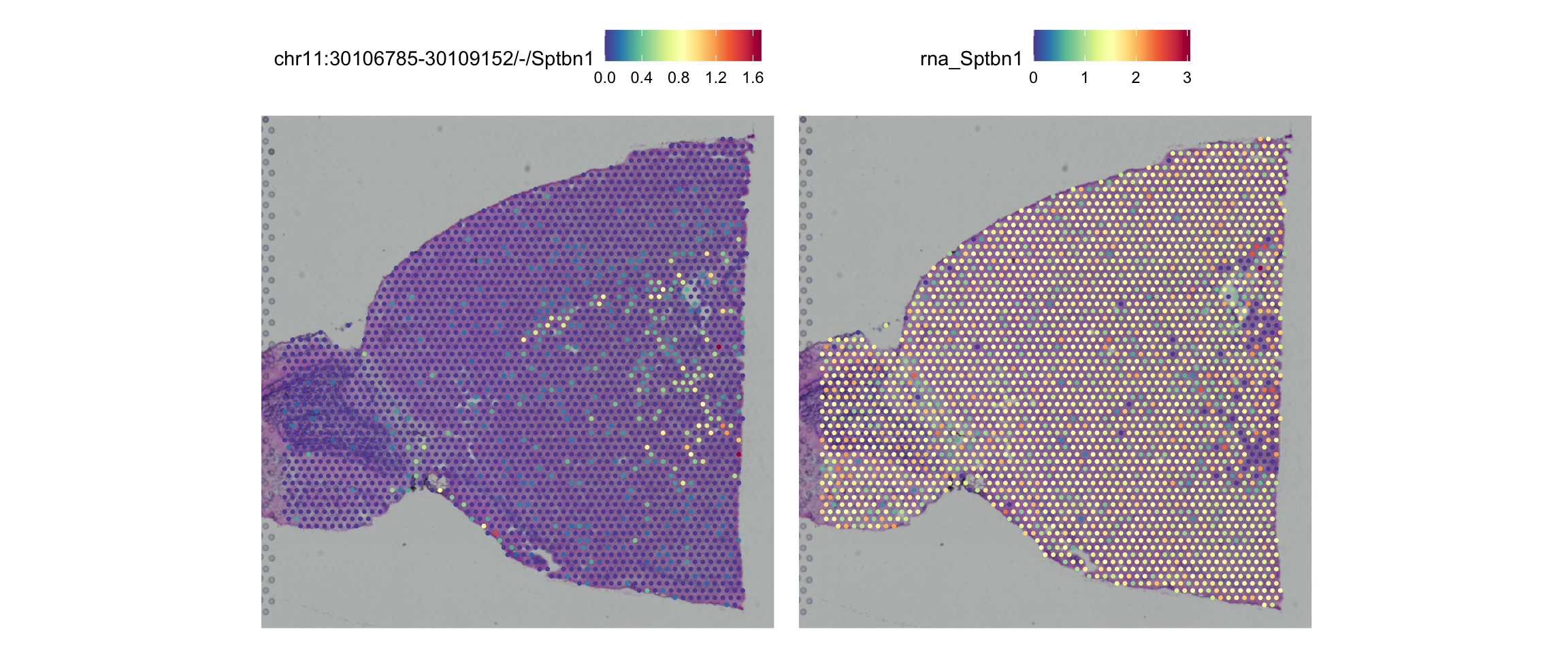
chr11:30106785-30109152/-/Sptbn1 chr11 30106785 30109152 Sptbn1 -
chr11:53541475-53541898/+/Sept8 chr11 53541475 53541898 Sept8 +
chr11:53541475-53541847/+/Sept8 chr11 53541475 53541847 Sept8 +
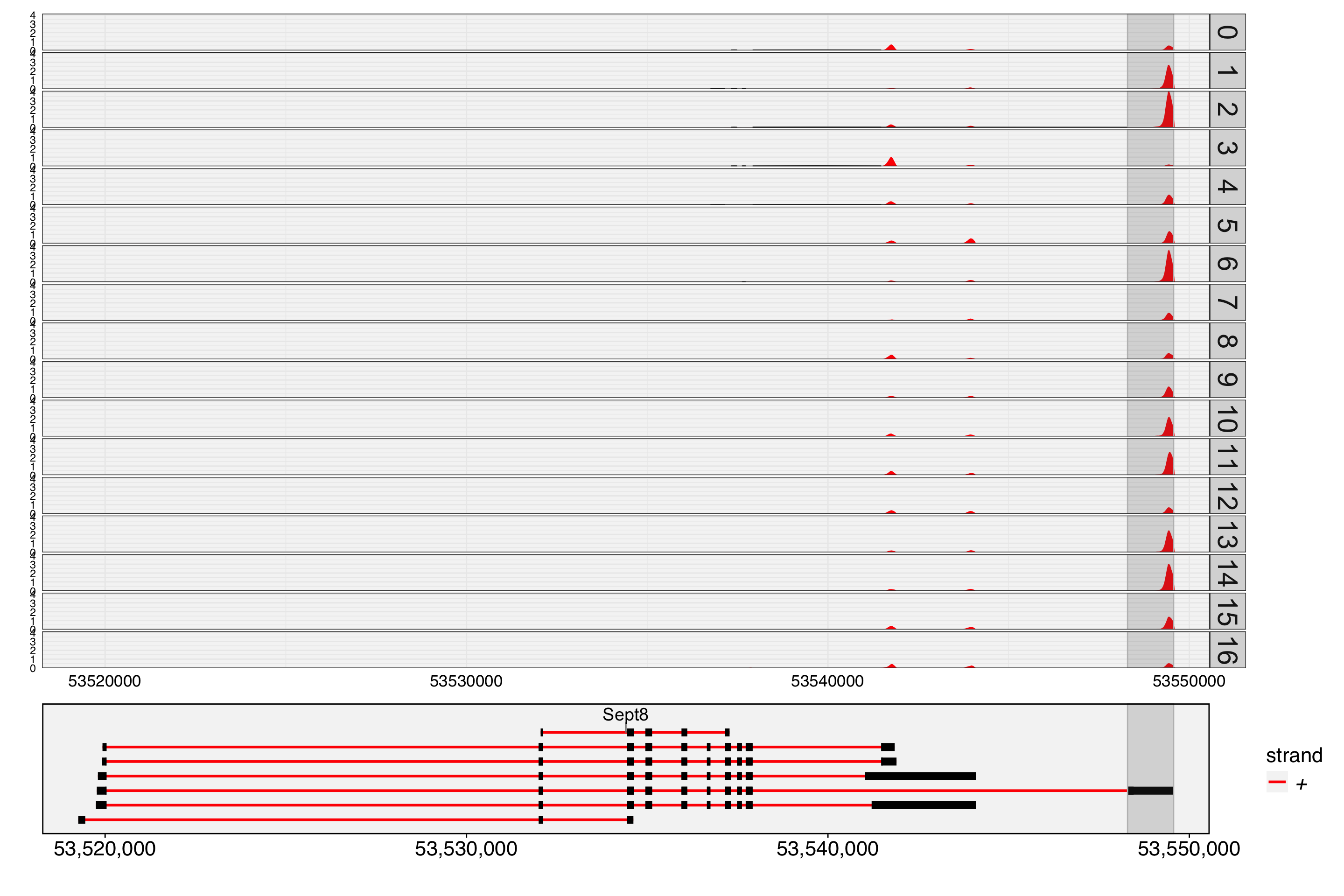
chr11:53548291-53549565/+/Sept8 chr11 53548291 53549565 Sept8 +
chr11:53541210-53544095/+/Sept8 chr11 53541210 53544095 Sept8 +
chr11:53541030-53544096/+/Sept8 chr11 53541030 53544096 Sept8 +
chr12:111806315-111806775/+/Klc1 chr12 111806315 111806775 Klc1 +
chr12:111806315-111806727/+/Klc1 chr12 111806315 111806727 Klc1 +
chr12:111806315-111806711/+/Klc1 chr12 111806315 111806711 Klc1 +
chr12:111806315-111806726/+/Klc1 chr12 111806315 111806726 Klc1 +
chr13:59128168-59133970/+/Ntrk2 chr13 59128168 59133970 Ntrk2 +
chr14:20507671-20508721/-/Ppp3cb chr14 20507671 20508721 Ppp3cb -
chr17:25717528-25717581/+/Gng13 chr17 25717528 25717581 Gng13 +
chr4:117154638-117155181/-/Rps8 chr4 117154638 117155181 Rps8 -
chr4:117154391-117154813/-/Rps8 chr4 117154391 117154813 Rps8 -
chr4:117154510-117154813/-/Rps8 chr4 117154510 117154813 Rps8 -
chr4:117155008-117155107/-/Rps8 chr4 117155008 117155107 Rps8 -
chr4:117155008-117155677/-/Rps8 chr4 117155008 117155677 Rps8 -
chr5:121205406-121205663/+/Rpl6 chr5 121205406 121205663 Rpl6 +
chr5:121205406-121206202/+/Rpl6 chr5 121205406 121206202 Rpl6 +
chr7:27180169-27180279/-/Gm21983 chr7 27180169 27180279 Gm21983 -
chr7:27180700-27180833/-/Gm21983 chr7 27180700 27180833 Gm21983 -
chr7:45131188-45131566/-/Flt3l chr7 45131188 45131566 Flt3l -
chr7:45131188-45131431/-/Flt3l chr7 45131188 45131431 Flt3l -
chrX:163926903-163929546/+/Ap1s2 chrX 163926903 163929546 Ap1s2 +
moransi.pval moransi autocorr.variable
chr11:30106785-30109152/-/Sptbn1 0.000000e+00 0.15168101 TRUE
chr11:53541475-53541898/+/Sept8 0.000000e+00 0.42945787 TRUE
chr11:53541475-53541847/+/Sept8 0.000000e+00 0.35888116 TRUE
chr11:53548291-53549565/+/Sept8 0.000000e+00 0.43158740 TRUE
chr11:53541210-53544095/+/Sept8 0.000000e+00 0.40952155 TRUE
chr11:53541030-53544096/+/Sept8 0.000000e+00 0.40944155 TRUE
chr12:111806315-111806775/+/Klc1 0.000000e+00 0.10015770 TRUE
chr12:111806315-111806727/+/Klc1 0.000000e+00 0.09999863 TRUE
chr12:111806315-111806711/+/Klc1 0.000000e+00 0.09420650 TRUE
chr12:111806315-111806726/+/Klc1 0.000000e+00 0.09698996 TRUE
chr13:59128168-59133970/+/Ntrk2 0.000000e+00 0.32438027 TRUE
chr14:20507671-20508721/-/Ppp3cb 0.000000e+00 0.06726973 TRUE
chr17:25717528-25717581/+/Gng13 0.000000e+00 0.16337588 TRUE
chr4:117154638-117155181/-/Rps8 0.000000e+00 0.21340388 TRUE
chr4:117154391-117154813/-/Rps8 0.000000e+00 0.29275023 TRUE
chr4:117154510-117154813/-/Rps8 0.000000e+00 0.29520866 TRUE
chr4:117155008-117155107/-/Rps8 0.000000e+00 0.21347766 TRUE
chr4:117155008-117155677/-/Rps8 0.000000e+00 0.20857395 TRUE
chr5:121205406-121205663/+/Rpl6 1.476043e-309 0.06564243 TRUE
chr5:121205406-121206202/+/Rpl6 1.483145e-315 0.06627969 TRUE
chr7:27180169-27180279/-/Gm21983 0.000000e+00 0.11385203 TRUE
chr7:27180700-27180833/-/Gm21983 0.000000e+00 0.13438134 TRUE
chr7:45131188-45131566/-/Flt3l 1.183112e-115 0.03978920 TRUE
chr7:45131188-45131431/-/Flt3l 9.910321e-99 0.03667860 TRUE
chrX:163926903-163929546/+/Ap1s2 0.000000e+00 0.14474266 TRUE
gene_name.D gene_name.t gene_name.pval
chr11:30106785-30109152/-/Sptbn1 0.4746839 -43.89401 5.263807e-67
chr11:53541475-53541898/+/Sept8 0.4048398 -25.74381 5.834218e-46
chr11:53541475-53541847/+/Sept8 0.3656638 -22.48278 5.567885e-41
chr11:53548291-53549565/+/Sept8 0.4780168 -26.93290 1.160903e-47
chr11:53541210-53544095/+/Sept8 0.3912296 -24.47323 4.461505e-44
chr11:53541030-53544096/+/Sept8 0.3913941 -24.46688 4.561207e-44
chr12:111806315-111806775/+/Klc1 0.4504790 -34.17159 6.621251e-57
chr12:111806315-111806727/+/Klc1 0.4505177 -34.03884 9.447059e-57
chr12:111806315-111806711/+/Klc1 0.4413015 -32.84301 2.449418e-55
chr12:111806315-111806726/+/Klc1 0.4444515 -33.46269 4.478730e-56
chr13:59128168-59133970/+/Ntrk2 0.4148224 -24.83806 1.263263e-44
chr14:20507671-20508721/-/Ppp3cb 0.3496615 -22.53752 4.550498e-41
chr17:25717528-25717581/+/Gng13 0.4628582 -28.12830 2.578332e-49
chr4:117154638-117155181/-/Rps8 0.2055551 -22.67264 2.769229e-41
chr4:117154391-117154813/-/Rps8 0.2711817 -34.15592 6.904540e-57
chr4:117154510-117154813/-/Rps8 0.2710378 -34.78865 1.288374e-57
chr4:117155008-117155107/-/Rps8 0.2653470 -30.72746 9.963707e-53
chr4:117155008-117155677/-/Rps8 0.2590419 -28.16428 2.303631e-49
chr5:121205406-121205663/+/Rpl6 0.2384405 -23.27607 3.087837e-42
chr5:121205406-121206202/+/Rpl6 0.2388061 -24.60967 2.778846e-44
chr7:27180169-27180279/-/Gm21983 0.3214447 -22.61010 3.484121e-41
chr7:27180700-27180833/-/Gm21983 0.3414768 -22.39562 7.682987e-41
chr7:45131188-45131566/-/Flt3l 0.2651879 -22.54243 4.469064e-41
chr7:45131188-45131431/-/Flt3l 0.2518174 -22.66769 2.820021e-41
chrX:163926903-163929546/+/Ap1s2 0.3696098 -24.50636 3.976343e-44
gene_name.padj
chr11:30106785-30109152/-/Sptbn1 2.008300e-62
chr11:53541475-53541898/+/Sept8 1.854941e-42
chr11:53541475-53541847/+/Sept8 8.851314e-38
chr11:53548291-53549565/+/Sept8 4.026538e-44
chr11:53541210-53544095/+/Sept8 1.023669e-40
chr11:53541030-53544096/+/Sept8 1.023669e-40
chr12:111806315-111806775/+/Klc1 6.585722e-53
chr12:111806315-111806727/+/Klc1 7.208673e-53
chr12:111806315-111806711/+/Klc1 1.335038e-51
chr12:111806315-111806726/+/Klc1 2.847950e-52
chr13:59128168-59133970/+/Ntrk2 3.707483e-41
chr14:20507671-20508721/-/Ppp3cb 7.548485e-38
chr17:25717528-25717581/+/Gng13 9.837110e-46
chr4:117154638-117155181/-/Rps8 5.379613e-38
chr4:117154391-117154813/-/Rps8 6.585722e-53
chr4:117154510-117154813/-/Rps8 2.457766e-53
chr4:117155008-117155107/-/Rps8 4.751816e-49
chr4:117155008-117155677/-/Rps8 9.765604e-46
chr5:121205406-121205663/+/Rpl6 6.545014e-39
chr5:121205406-121206202/+/Rpl6 7.572952e-41
chr7:27180169-27180279/-/Gm21983 6.329984e-38
chr7:27180700-27180833/-/Gm21983 1.172516e-37
chr7:45131188-45131566/-/Flt3l 7.548485e-38
chr7:45131188-45131431/-/Flt3l 5.379613e-38
chrX:163926903-163929546/+/Ap1s2 1.011396e-40